Caixia Gao team Unveil AI-Powered Universal Strategy for Protein Engineering
Dr.Caixia Gao team has developed a groundbreaking method that could transform the field of protein engineering. The new approach, called AI-informed Constraints for protein Engineering (AiCE), enables rapid and efficient protein evolution by integrating structural and evolutionary constraints into a generic inverse folding model—without the need to train specialized artificial intelligence (AI) models.This study was published on Cell on July 7 (10.1016/j.cell.2025.06.014), addressing many challenges associated with traditional protein engineering techniques.
The ideal protein engineering strategy would achieve optimal performance with minimal effort. However, the existing approaches are limited in cost, efficiency or scalability. The high computational demands of current AI-based protein engineering methods necessitate more accessible, user-friendly alternatives that preserve predictive accuracy and foster wider adoption by researchers.
In this study, the researchers started with the development of AiCEsingle, a module designed to predict high-fitness (HF) single amino acid substitutions. It boosted prediction accuracy by extensively sampling inverse folding models—AI models that generate compatible amino acid sequences based on protein 3D structures—while incorporating structural constraints.
For further validation, the researchers benchmarked AiCEsingle on 60 deep mutational scanning (DMS) datasets, demonstrating its superior performance over other AI-based methods by 36%–90%. Its effectiveness was also validated for complex proteins and protein–nucleic acid complexes. Notably, incorporating structural constraints alone achieved a 37% improvement in accuracy. To address the challenge of negative epistatic interactions in combinatorial mutations, the AiCEmulti module was developed. This advanced package enables accurate prediction of multiple high-fitness mutations with minimal computational cost, significantly expanding the tool's versatility and practical utility.
Utilizing the AiCE framework, the research team engineered eight distinct proteins spanning multiple structural folds and functional classes, including deaminases,nuclear localization signals, nucleases, and reverse transcriptases. These engineered proteins were further successfully applied in next-generation base editors for precision medicine and molecular breeding. Specifically, enABE8e is a cytosine base editor with a ~50% narrower editing window; enSdd6-CBE is an adenine base editor with 1.3× higher fidelity; and enDdd1-DdCBE is a mitochondrial base editor showing a 13× increase in activity.
AiCE established a streamlined, efficient and universally applicable paradigm for protein engineering. By leveraging existing AI architectures, this framework illuminates a transformative trajectory for the field while conferring enhanced mechanistic interpretability to AI-driven protein redesign.
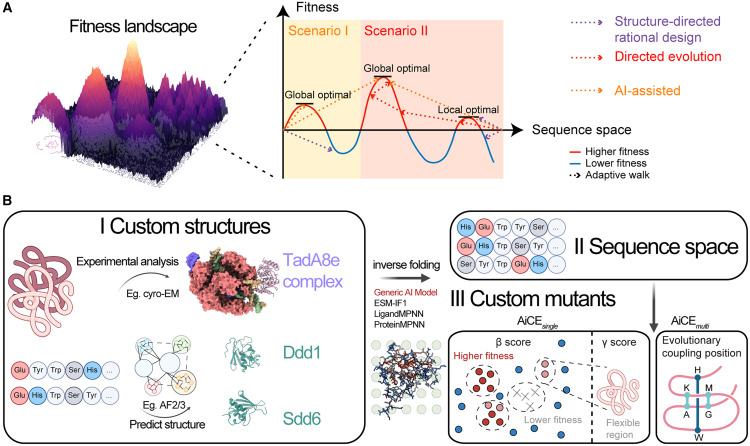
Figure 1. AiCE as an AI-informed approach for protein engineering
File Download:
